Genome-wide Screening in Diverse Bacteria with SLALOM and Mobile CRISPRi
Published on: July 28, 2022
Updated on: July 28, 2022
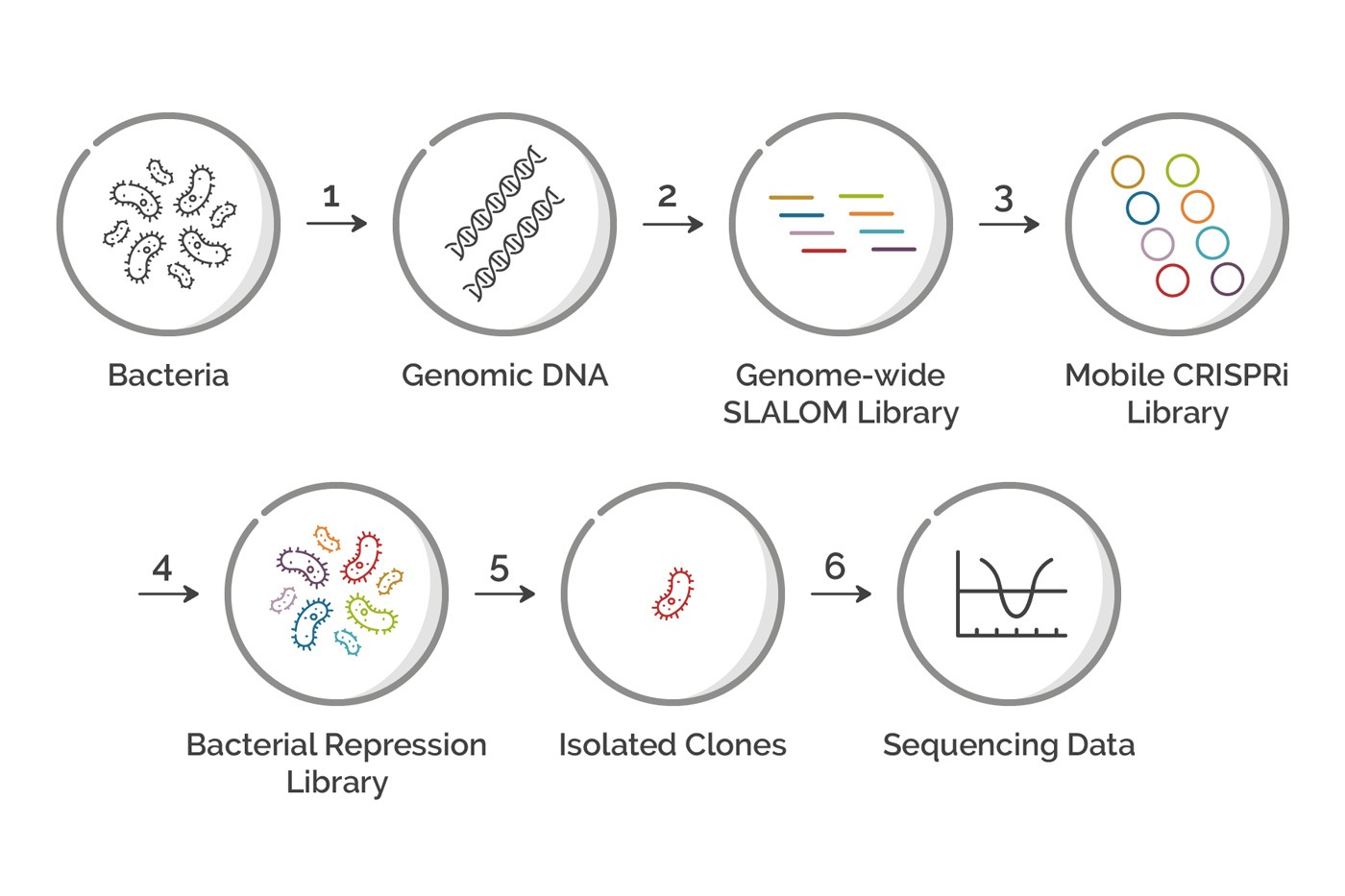
Figure 1: Flowchart showing each step in a Genome-wide CRIPSRi Screen in Bacteria. Each step in the process is described in detail in the article below.
Why Screen in Bacteria?
There are likely over a trillion different species of bacteria on earth. These tiny organisms are able to thrive in the most remote places thanks to an enormous arsenal of enzymes that have evolved over the last few billion years. For instance, some bacteria known as lithophiles are able to extract energy from rocks on the bottom of the ocean. Others, called acidophiles, produce durable proteins that keep them comfortable in extremely acidic environments. The breadth of molecular function encoded in the genomes of bacteria make them a massive repository of useful enzymes that have the potential to be used in everything from food preparation to biopharmaceuticals. Until recently, genetic techniques to discover these enzymes were not available in the vast majority of bacteria. However, it is now possible to explore unstudied genomes with tools like Mobile CRISPRi and SLALOM. In this article we briefly describe these two technologies and walk through each step of a genome-wide CRISPRi screen for genes including those coding for undiscovered enzymes.
Genetic Tools
Before walking through the steps of a genome-wide screen in bacteria, it is important to have a basic understanding of the technologies involved in the process and how they work together. Here is a quick overview of the technologies we will be discussing.
CRISPR interference (CRISPRi) is a technique to repress expression of a gene. In bacteria, when a nuclease deficient Cas9 (dCas9) and single guide RNA (sgRNA) are expressed in a cell, the Cas9-sgRNA complex will bind to the non-template strand of the gene targeted by the sgRNA and physically block extension of the RNA polymerase. This causes the polymerase to dissociate from the DNA, effectively reducing the number of functional transcripts of that gene. This is an easy way to repress transcription of any gene made by the bacteria. However, it can be difficult to deliver a construct carrying Cas9 and the sgRNA into most bacteria other than well established laboratory strains.
Mobile CRISPRi is a system that uses bacterial conjugation and Tn7 transposition to integrate a construct carrying dCas9 and an sgRNA into the chromosome of a bacterium. It is compatible with a wide range of bacteria, including both gram positive and gram negative bacteria. A suite of modular Mobile CRISPRi plasmids have been developed that make it possible to switch out promoters and antibiotic resistance genes for specific use cases. While the system can be used to repress a single gene, it can also be used to repress all genes in a genome in a single experiment. These genome-wide studies require the synthesis of a pool of thousands of short DNA oligos encoding unique sgRNAs that target each individual gene. Genome-wide sgRNA libraries can be expensive and can take time to make.
SLALOM is a method to quickly and cost effectively generate sgRNA libraries directly from DNA. Traditionally, sgRNA library synthesis has been accomplished by sending a text file containing thousands of unique targeting sequences to a company capable of manufacturing microarrays. Clusters of each oligo are synthesized on a glass slide and cleaved off to create a pool of oligos. SLALOM takes a different approach. The method involves a short series of steps that enzymatically converts the DNA into an sgRNA library. It works on the principle of extracting valid targeting sequences from the DNA that are adjacent to PAM motifs. Eliminating the need to bioinformatically design the library and to have it chemically synthesized significantly reduces the cost, effort, and time needed to make a library. Libraries constructed using SLALOM can be adapted for compatibility with all CRISPR based systems, including Mobile CRISPRi.
Steps in a Genome-wide CRIPSRi Screen
By using Mobile CRISPRi and a genome-wide library generated with SLALOM it’s possible to quickly screen for genes in diverse bacteria. Here we outline the details of each step involved in a genome-wide CRISPRi screen using these technologies.
Step 1 - Isolate Genomic DNA from the Bacteria
Genomic DNA from the bacteria you are working with must be purified before a genome-wide sgNRA library can be generated with SLALOM. Most standard microbial genomic DNA purification kits such as the DNeasy PowerLyzer Microbial Kit (QIAGEN) will provide high quality DNA. The resulting sgRNA library will contain targeting sequences from the source of DNA that is used.
Step 2 - Synthesize a Genome-wide sgRNA Library with SLALOM
The purified genomic DNA can be enzymatically converted into a Genome-wide sgRNA library using the SLALOM 1.0™ sgRNA Library Synthesis Kit (Pioneer Biolabs). The entire procedure takes less than a day and can be performed on the bench using basic lab equipment. A manual with the complete protocol can be downloaded here, and kits can be found here. The kit produces high-quality, variant-matched sgRNA libraries targeting the genome of the species or strain of interest. For E. coli MG1655, the kit produces a library composed of about 46,000 unique sgRNAs, which is on average about 2 sgRNAs (both forward and reverse strands) every 200 bp of the genome.
Step 3 - Clone the sgRNA Library into a Mobile CRISPRi Plasmid
Several Mobile CRISPRi plasmids are available for cloning the genome-wide SLALOM library (see “Suggested Mobile CRISPRi Plasmids” below). These plasmids include the coding region for Cas9 and a cloning site for the sgRNAs. Digesting the plasmid and the library with the restriction enzyme BasI creates compatible sticky ends that position the library in the correct orientation after ligation. A molar ratio of 3:1 of the library to plasmid is recommended as a starting point for optimizing the ligation if needed.
Mobile CRISPRi plasmids have an R6K origin of replication, which means that they only replicate in cells expressing the pi protein. For cloning large libraries it is recommended that you electroporate the ligated plasmids into a highly efficient electrocompetent strain of E. coli such as the TransforMax EC100D pir+ Electrocompetent E. coli from Lucigen (now part of LGC Biosearch Technologies). Transformed cells are plated so that colonies can be counted. For initial library cloning, it’s important to have 100-500 colony-forming units (cfu) per sgRNA in the library to maintain even representation of the sgRNAs. Alternatively, libraries have been successfully cloned in liquid culture by ensuring the culture stays below OD600 of 0.4 to prevent competition. For liquid cultures, the number of cfu can be estimated by plating a few microliters of the culture broth. In either situation, the cells are then harvested and the cloned Mobile CRISPRi plasmid library can then be purified using a standard midi-prep kit.
Step 4 - Chromosomal Integration of the Mobile CRISPRi Library
The purified Mobile CRISPRi library is then integrated into the chromosome of the bacteria you are working with. This is accomplished by transforming the bacteria with both the cloned library and a helper plasmid expressing four of the five Tn7 transposase genes tnsABCD. Expression of these 4 genes results in site specific integration of the mobile CRISPRi plasmid at the attTn7 site located 25 bp downstream of the highly conserved glmS gene. Transformation can be carried out using electroporation, if the bacteria can be made electrocompetent, or by triparental mating. Triparental mating requires that the Mobile CRISPRi library and the helper plasmid are transferred into a mating strain expressing the conjugation machinery. Integration is selected for by growing on plates or media containing the correct antibiotic but lacking diaminopimelic acid (DAP). The established bacterial repression library can be used in many different screening setups depending on the phenotype being studied. Repression of each gene in the library is induced by adding IPTG to the culture.
Steps 5 & 6 - Isolate Clones and Identify Genes
There are several ways to isolate clones with the phenotype of interest and to identify the genes associated with the phenotype. To illustrate some of these techniques, We will look at an example of identifying enzyme E which is capable of metabolizing substrate S.
The Plating method
Suppose the bacteria you are working with turn red when they metabolize substrate S, otherwise they appear white. This visual indicator of metabolism could be used to screen for the gene encoding enzyme E. The bacterial repression library is plated on large plates containing substrate S and IPTG to isolate single colonies and induce expression of the Cas9 and the sgRNAs. Most colonies will appear red, but a few will be white, indicating that enzyme E is being repressed in these cells. One of these colonies is picked and the targeting region of the sgRNA in that cell is Sanger sequenced and aligned back to the genome to determine which gene is being repressed. This gene is a candidate for encoding enzyme E. Additional colonies can be picked and sequenced. Identification of a few different sgRNAs targeting the same gene increases the confidence that the gene encodes enzyme E.
The Sorting Method
After sequencing some of the colonies on the large plates, it becomes apparent that there are actually multiple enzymes involved in the metabolism of substrate S. If this is the case, picking and sequencing individual colonies could quickly become unwieldy. Fortunately, the color difference between the cells that metabolize substrate S and those that do not can be used to sort the cells into two subpopulations using Fluorescence-activated Cell Sorting (FACS). The bacterial repression library is grown in liquid broth containing IPTG and the cells are sorted based on color. Genomic DNA from the white cells is then purified and the sgRNA sequences are PCR amplified and sent off for high through-put sequencing. The genes are then aligned back to the genome to determine candidate genes involved in the metabolism of substrate S.
The Enrichment Method
There are many situations where there is no good visual indicator of metabolism. However, it is often possible to apply a selective pressure on a population of cells to increase the relative abundance of cells capable of metabolizing a particular substrate. For example, it might turn out that cells capable of metabolizing substrate S grow faster than those that do not. In this case, two different flasks of the bacterial repression library could be grown for 15 to 20 generations, one with IPTG induction and one without. The bacteria in each flask are then pelleted and the genomic DNA is isolated. The sgRNA sequences from each sample are amplified and the two samples are sent for high-throughput sequencing. The uninduced control serves as a baseline for comparison in determining the relative representation of each gene. Because cells that are incapable of metabolizing substrate S grow slower, the sgRNAs from these cells should be underrepresented in the IPTG induced population. Genes that have a decreased fold change are candidates for genes involved in metabolism of substrate S. Replicates are needed to help avoid false positive and false negative results and to calculate statistical significance.
Summary
Useful enzymes produced by many species of bacteria on the earth have not been identified because genetic tools have not been available. Tools like SLALOM and Mobile CRISPRi make it possible to screen for genes in diverse bacteria. SLALOM can be used to generate a genome-wide sgRNA library at a fraction of the cost of traditional library synthesis. Using the method it’s possible to finish a genome-wide screen in 1-2 weeks. There are many different ways that a screen can be used to study a gene or a group of genes involved in a process such as the metabolism of a particular substrate.
Have additional questions or want to talk about designing a genome-wide CRISPRi screen? Please feel free to reach out to us to schedule a free consulting appointment at: support@pioneerbiolabs.com
Suggested Materials
Microbial Genomic DNA Extraction
DNeasy PowerLyzer Microbial Kit (QIAGEN)
Genome-wide sgRNA Library Synthesis
SLALOM 1.0 ™ sgRNA Library Synthesis Kit - Mobile CRISPRi version (Pioneer Biolabs)
Mobile CRISPRi Plasmids
pJMP1039 - Expresses Tn7 transposase
pJMP2834 - Test plasmid, encodes sfGFP and sfGFP targeting sgRNA (amp, kan)
pJMP2846 - For cloning new sgRNAs (amp, kan)
References
1. Yates JD, Russell RC, Barton NJ, Yost HJ, Hill JT. A simple and rapid method for enzymatic synthesis of CRISPR-Cas9 sgRNA libraries. Nucleic Acids Res. 2021;49(22):e131.
2. Peters JM, Koo BM, Patino R, et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat Microbiol. 2019;4(2):244-250.
3. Mitra R, McKenzie GJ, Yi L, Lee CA, Craig NL. Characterization of the TnsD-attTn7 complex that promotes site-specific insertion of Tn7. Mob DNA. 2010;1(1):18.